With one in three people affected at some point in their lives cancer is a condition we’re all acquainted with. However, one cancer that may not be familiar is oesophageal aka cancer of the food pipe (the tube used to carry food from our mouths to our stomachs). Over the last quarter of a century the incidence of oesophageal cancer has been on the rise, particularly in the UK, making it now the 8th most common cancer worldwide.
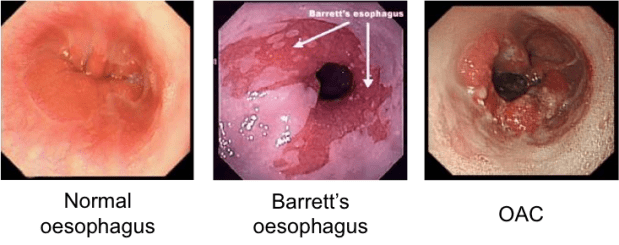
As we know cancer occurs as a result of uncontrolled proliferation (growth) of abnormal cells, in this case in the oesophagus. Oesophageal cancer commonly takes one of two forms namely Squamous Cell Carcinoma (SCC) or Oesophageal Adenocarcinoma (OAC), with recent years seeing a rise of the latter. One common risk factor for OAC is thought to be a condition known as Barrett’s Oesophagus, which develops in response to prolonged exposure to stomach acid and bile due to gastro-oesophageal reflux disease (GORD). This exposure can trigger changes in the types of cells found in the lining of the oesophagus. However, whether Barrett’s Oesophagus is always required for disease progression has yet to be confirmed. Also missing from our understanding is any real clear molecular insight into the changes occurring between normal oesophageal cells and resulting carcinomas. Thus it is no wonder that OAC is associated with a poor prognosis and five year survival rate of just 14%.
ATAC-seq – the ultimate technique?
Traditional studies investigating cancer development focus on that one gene / set of genes believed to be causing all the problems. However, in recent years, with the development of many next-generation sequencing techniques, this highly biased approach has been called into question. Over in the Sharrock’s lab here in Manchester, 3rd year PhD student Connor Rogerson is overcoming this bias using a method known as ATAC-seq (Assay for Transposase-Accessible Chromatin Sequencing – which is a bit of a mouthful)! ATAC-seq is a tool that allows researchers to identify regions of open chromatin (accessible DNA) and view these regions as peaks in a browser. Accessible DNA can often be mapped to regions of the genome that are highly active and temporally bound by molecules (known as transcription factors) that help switch genes on. Overall ATAC-seq helps to reveal the unique molecular signature of the cells / sample in question.
Biopsies, bioinformatics and biomarkers
Looking in both normal and oesophageal cancer samples from patient biopsies, Connor identified numerous regions showing differences in DNA accessibility between healthy and cancerous cells. Rather than looking at what genes became more accessible in the cancer cells, Connor switched the focus to what was turning these genes on, i.e. what transcription factors were associated with these cancer-cell accessible regions. Transcription factors bind to unique sequences of DNA known as motifs. By working out which motifs are most prevalent in cancer-cell accessible regions we can work out which transcription factors are in abundance and potentially driving the molecular changes associated with OAC. From this, transcription factors HNF4 and GATA6 were identified as possible gene activating factors in oesophageal cancer. Interestingly these factors were also present in abundance in Barrett’s Oesophagus (a potential early indicator of OAC) highlighting their potential as early stage biomarkers.
So what’s next?
With the jury still out on what the driving factor for OAC is, and the role of HNF4 and GATA6 found in both Barrett’s and OAC samples, the search is now on to uncover which genes are being turned on / off between these two possible stages. With presentation of OAC often not until the late stage, work performed by Connor and new team members Sam Ogden and Will Bleaney could help lead to the identification of early molecular changes driving this transition, key to improved prognosis and survival.
Discover more from Research Hive
Subscribe to get the latest posts sent to your email.
One thought on “Researcher Spotlight: Connor Rogerson”